Abstract
Achieving a feasible generation of realistic datasets for computer vision-based ML models involves explorative methods beyond the basic techniques of image augmentation like flip, rotate, random translation, and mirror-symmetrical replicas of images. However, addressing the suitability and generation of almost real-time datasets need help from Deep Learning techniques. This report describes an implementation of a Generative Adversarial Network-based augmentation technique called LeafGAN, adapted from an Image-to-Image translation algorithm CycleGAN. This method is capable of augmenting leaf disease images from normal leaf images with diverse backgrounds.
Introduction
Deep learning has become a standard tool for many applications in computer vision, since the accomplishment of model generalization from computing massive datasets. However, in reality, datasets are hard to combine for new applications and environments as diverse data collection consumes time, cost and effort. Data augmentation provides a good source of diverse data for training models, without the requirement of collecting new data samples. With the basic image augmentation techniques like flipping, random noise, crop and rotation, though it is possible to generate data, the data shift challenges remain the same. Hence, one cannot expect the models to be robust against data shifts and data corruption.
Value of augmentation
Typically, augmentation techniques generate value for many real-time detection deployments. As per Gartner’s AI business value forecast, decision support/augmentation will surpass all the other types of AI initiatives and will account for 44% of the global AI-derived business value by 2030. Also according to 2021 expectations of Gartner, Artificial intelligence (AI) augmentation is expected to create $2.9 trillion of business value and 6.2 billion hours of workers’ productivity globally, resulting in the fact that Image augmentation will have a major contribution to Computer Vision based AI solutions.
Plant disease Diagnosis
Automated diagnosis of plant disease is one of the most active research fields in agriculture which uses the techniques of computer vision and deep learning. Many deep learning-based techniques for the automated diagnosis of plant diseases have been reported with an aim of supporting farmers and reducing loss in terms of plant productivity.
Challenges include the requirement of
- A huge number of training images
- Labelled disease datasets requiring domain knowledge
- Generating gold standard (ground truth) datasets
Other bottlenecks include
- A strictly controlled and isolated environment should be used to avoid contamination
- Class imbalance
- Overfitting issues as the image features are tiny
- Unknown/new environment (background) is a bottleneck for accuracy
Though collecting healthy images are easy, the background diversity of disease images tends to be limited due to ambient conditions such as weather, temperature and vector-borne insects.
Disease classification models are generally biased towards classes with more samples and higher variation. Overfitting is a serious concern as the target object is of small size and the clues for diagnosis may be just a dot or tiny wrinkles. In general, a deep classifier such as a CNN tends to capture the Image features (brightness/colour) of a large area. Finally when evaluating the classifier models,accuracy tends to be higher with the trained and evaluation datasets but ultimately fails many times with new/unknown environments.
Implementation
In this report, we will explore and implement an advanced data augmentation technique applied for Leaf Image generation known as LFLSeg which is a part of LeafGAN based on another image-to-image translation method CycleGAN.
Clone the GitHub repository using the command
git clone https://github.com/IyatomiLab/LeafGAN
Install the dependencies using pip
pip install torch>=1.4.0
pip install torchvision>=0.5.0
pip install dominate>=2.4.0
pip install visdom>=0.1.8.8
pip install requests>=2.23.0
pip install opencv_python_headless>=4.2.0.34
Collect the dataset
The initiation of the partial leaf class is LFLSeg's main concept wherein full leaf photos are used to make partial leaf images as we crop 9 patches from a single complete leaf image to get 9 partial leaf photos, as shown below.

We can split the image into tiles of equal sizes using the image slicer package in python.
Sample snippet of code
import os
path = 'path to the images to be sliced’
files = os.listdir()
#
for file in files:
import image_slicer
image_slicer.slice(file, 9)
Create a text file with training information with the following label:
Label 0 for full leaf, 1 for partial leaf, and 2 for non-leaf.
/path/to/full_leaf/full_leaf_1.JPG, 0
/path/to/full_leaf/full_leaf_2.png, 0
/path/to/full_leaf/full_leaf_3.jpg, 0
... ... ...
/path/to/partial_leaf/partial_leaf_1.JPG, 1
/path/to/partial_leaf/partial_leaf_2.jpg, 1
/path/to/partial_leaf/partial_leaf_3.png, 1
... ... ...
/path/to/non_leaf/non_leaf_1.png, 2
/path/to/non_leaf/non_leaf_2.jpg, 2
/path/to/non_leaf/non_leaf_3.JPG, 2
Train the model
To train the model using the python command
python train_LFLSeg.py --train /path/to/train_data.txt --test /path/to/train_data.txt --batch_size 20
Change the trained model path at line 91 of the leaf_gan_model.py file after training.
load_path = '/path/to/LFLSeg_resnet101.pth'
To obtain the GradCAM or the entire leaf picture mask. Execute the command below:
python infer_LFLSeg.py --input /path/to/sing_full_leaf_image
--segment --threshold
# --segment ( #if not given, segment flag will be False
#--threshold #value to get the masked image ([0.0, 1.0]),
Example
python infer_LFLSeg.py --input images/leaf_01.jpg --threshold 0.35 --segment
LFL-Seg Results
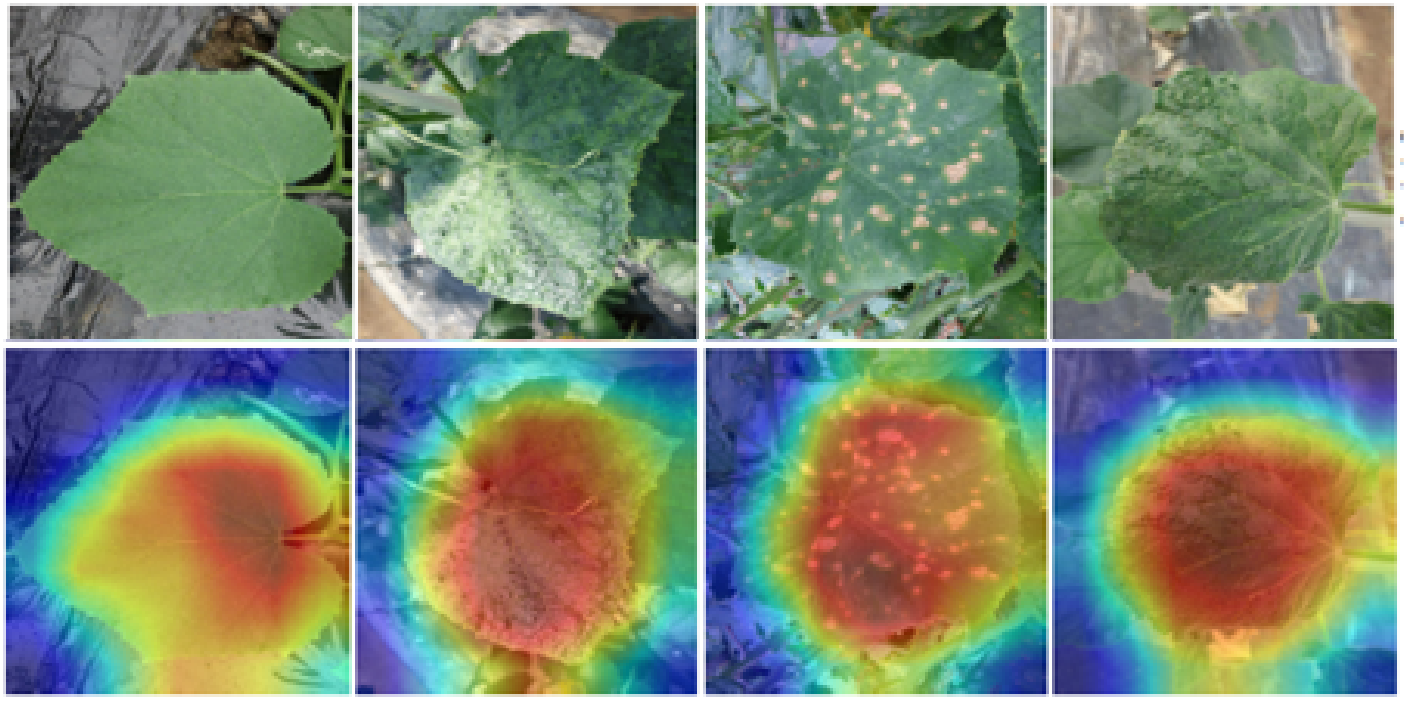
The heatmaps comparison of LFLSeg models trained with “partial leaf” images. The warmer the coloured region, the more it contributes to the final decision for a class (i.e., “full leaf” in this case).
Simple Augmentation
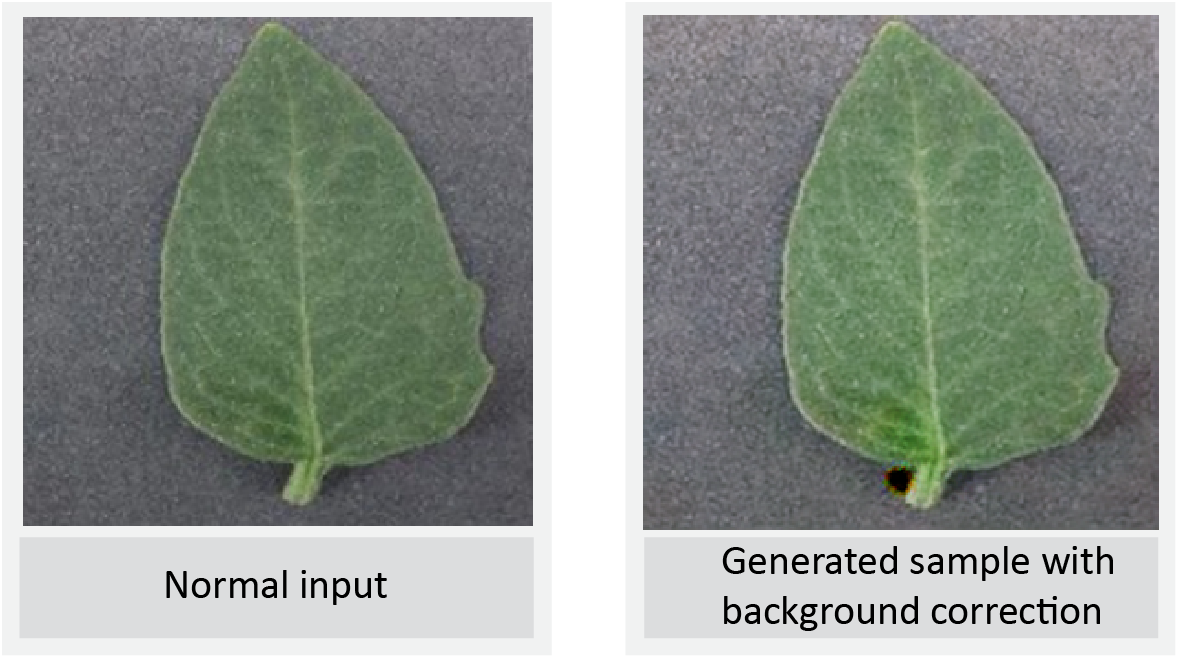
LeafGAN Output 1
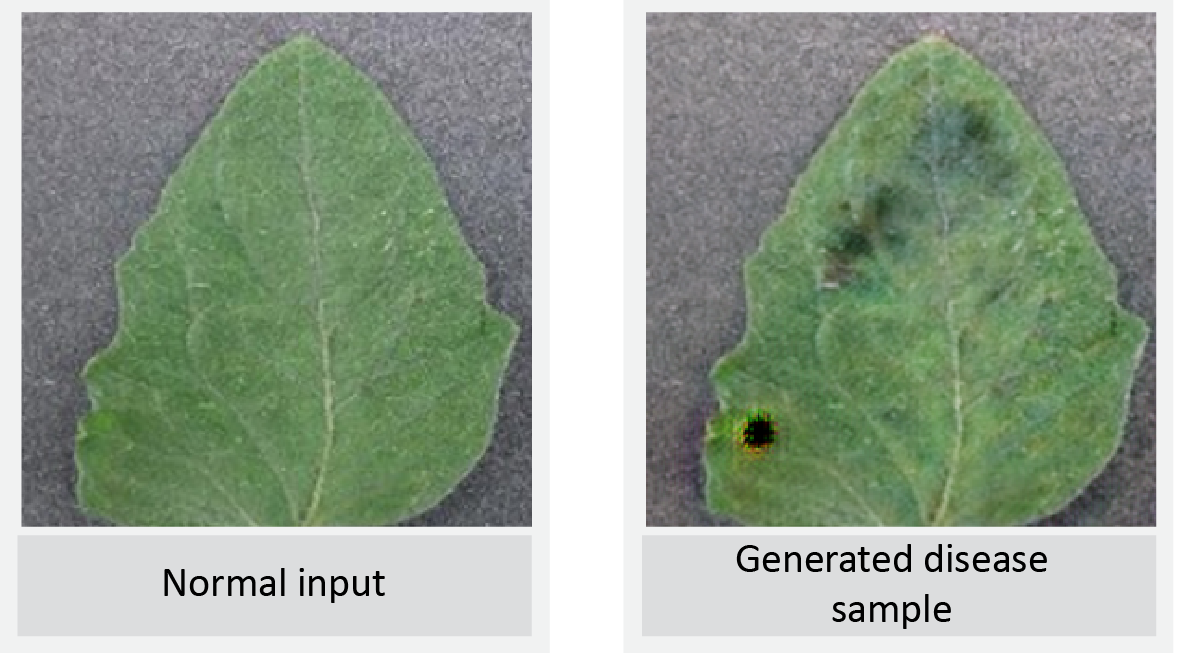
LeafGAN Output 2
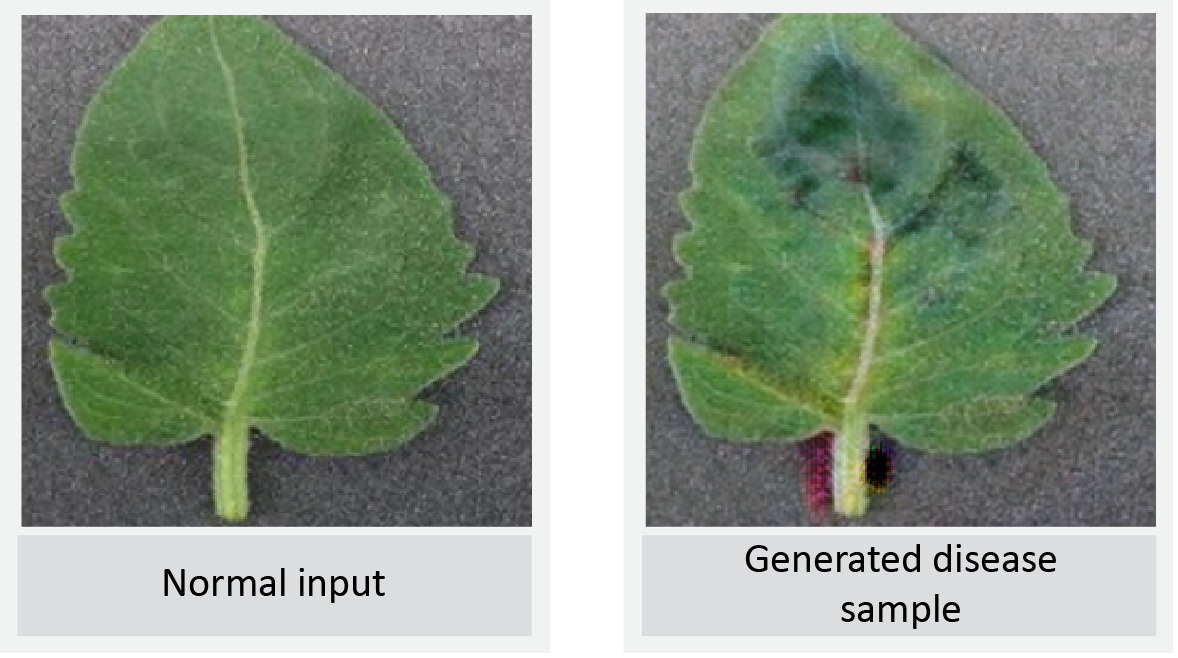
Conclusion
Data is becoming a tangible asset in the AI space and it becomes a bottleneck when the right data in context is hard to get. Augmenting and generating real-time dataset grows business assets and one can run manipulations on existing data. This allows data scientists to generate insights that were traditionally locked. In this article, we have discussed the implementation of data augmentation techniques related to plant-based leaf disease datasets.
References